|
 |
Articles |
| Coronavirus as a possible cause of severe acute respiratory syndrome |
J S M Peiris, S T Lai, L L M Poon, Y Guan, L Y C Yam, W Lim, J Nicholls,
W K S Yee, W W Yan, M T Cheung, V C C Cheng, K H Chan, D N C Tsang, R W
H Yung, T K Ng, K Y Yuen, and members of the SARS study group*
*Members listed at the end of paper
Department of Microbiology and Pathology, Queen Mary Hospital, University
of Hong Kong, Hong Kong (Prof J S M Peiris DPhil, L L M Poon DPhil, Y Guan PhD, J Nicholls FRCPA,
V C C Cheng MRCP, K H Chan PhD, Prof K Y Yuen FRCPath); Department of Medicine, Intensive Care and Pathology, Princess Margaret
Hospital, Hong Kong (S T Lai FRCP, W W Yan FRCP, T K Ng FRCPath); Government Virus Unit, Department of Health, Hong Kong (W Lim FRCPath); Department of Medicine and Pathology, Pamela Youde Nethersole Eastern
Hospital, Hong Kong (L Y C Yam FRCP, M T Cheung MRCP, R W H Yung FRCPath); Department of Medicine, Kwong Wah Hospital, Hong Kong (W K S Yee MRCP); and Department of Pathology, Queen Elizabeth Hospital, Hong Kong Special
Administrative Region, China (D N C Tsang FRCPath)
Correspondence to: Prof J S M Peiris, Department of Microbiology, University of Hong Kong,
Queen Mary Hospital, Pokfulam Road, Hong Kong, Special Administrative Region,
China (e-mail:malik@hkucc.hku.hk)Summary
Introduction
Methods
Results
Discussion
References
Background An outbreak of severe acute respiratory syndrome (SARS) has been reported
in Hong Kong. We investigated the viral cause and clinical presentation
among 50 patients.Methods We analysed case notes and microbiological findings for 50 patients with
SARS, representing more than five separate epidemiologically linked transmission
clusters. We defined the clinical presentation and risk factors associated
with severe disease and investigated the causal agents by chest radiography
and laboratory testing of nasopharyngeal aspirates and sera samples. We
compared the laboratory findings with those submitted for microbiological
investigation of other diseases from patients whose identity was masked.Findings Patients' age ranged from 23 to 74 years. Fever, chills, myalgia, and
cough were the most frequent complaints. When compared with chest radiographic
changes, respiratory symptoms and auscultatory findings were disproportionally
mild. Patients who were household contacts of other infected people and
had older age, lymphopenia, and liver dysfunction were associated with
severe disease. A virus belonging to the family Coronaviridae was isolated from two patients. By use of serological and reverse-transcriptase
PCR specific for this virus, 45 of 50 patients with SARS, but no controls,
had evidence of infection with this virus.Interpretation A coronavirus was isolated from patients with SARS that might be the primary
agent associated with this disease. Serological and molecular tests specific
for the virus permitted a definitive laboratory diagnosis to be made and
allowed further investigation to define whether other cofactors play a
part in disease progression.Lancet 2003; 361: 1319-25. Published online April 8, 2003http://image.thelancet.com/extras/03art3477web.pdf See Commentary

An outbreak of atypical pneumonia in Guangdong Province, People's Republic
of China, that has continued since November, 2002, is reported to have
affected 792 people and caused 31 deaths.1 In adjacent Hong Kong, surveillance of severe atypical pneumonia was heightened
in the public hospital network under the Hospital Authority of Hong Kong.
By the end of February, 2003, clusters of patients with pneumonia were
noted in Hong Kong, along with affected close contacts and health-care
workers. The disease did not respond to empirical antimicrobial treatment
for acute community-acquired typical or atypical pneumonia. Bacteriological
and virological pathogens known to cause pneumonia were not identified.
Thus, the new disorder was called severe acute respiratory syndrome (SARS).
Subsequently, SARS has spread worldwide to involve patients in North America,
Europe, and other Asian countries.1 We investigated patients in Hong Kong to try to identify the causal agent.

We included in the study 50 patients fitting a modified WHO definition
of SARS admitted to three acute regional hospitals in Hong Kong between
Feb 26 and March 26, 2003.2 Briefly, the case definition was fever of 38ºC or more, cough or
shortness of breath, new pulmonary infiltrates on chest radiography, and
a history of exposure to a patient with SARS or absence of response to
empirical antimicrobial coverage for typical and atypical pneumonia (ß
lactams and macrolides, fluoroquinolones, or tetracyclines).We collected nasopharyngeal aspirates and serum samples from all patients.
Paired acute and convalescent sera and faeces were available from some
patients. A lung-biopsy tissue sample from one patient was processed for
viral culture and reverse-transcriptase PCR (RT-PCR) and for routine histopathological
examination and electron microscopy. We used as controls nasopharyngeal
aspirates, and faeces and sera submitted for microbiological investigation
of other diseases from patients whose identities were masked.The SARS patients' medical records were reviewed retrospectively by the
attending physicians and clinical microbiologists. Routine haematological,
biochemical, and microbiological work-up was done, including bacterial
culture of blood and sputum, serology, and nasopharyngeal aspirates for
virology. The nasopharyngeal aspirate was assessed by rapid immunoflourescent
antigen detection for influenza A and B, parainfluenza types 1, 2, and
3, respiratory syncytial virus and adenovirus,3 and was cultured for conventional respiratory pathogens on Mardin Darby
Canine Kidney, LLC-Mk2, RDE, Hep-2 and MRC-5 cells.4 Subsequently, fetal rhesus kidney (FRhK-4) and A-549 cells were added
to the panel of cell lines used. RT-PCR for influenza A5 and human metapneumovirus was done directly on the clinical samples. The
degenerate primers used for human metapneumovirus were: first round 5'-AARGTSAATGCATCAGC-3'
and 5'-CAKATTYTG CTTATGCTTTC-3'; nested primers: 5'-ACACCTGT TACAATACCAGC-3'
and 5'-GACTTGAGTCCCA GCTCCA-3' (sequences using the International Union
of Pure Chemistry one-letter code). The size of the nested PCR product
was 201 bp. We used an ELISA for mycoplasma to screen cell cultures (Roche
Diagnostics, Indianapolis, IN, USA).Serology and detection of coronavirus After culture and genetic sequencing of a coronavirus from two patients,
we developed an RT-PCR to detect the coronavirus sequence from nasopharyngeal
aspiration samples. Total RNA from clinical samples was reverse transcribed
with random hexamers and cDNA was amplified with primers 5'-TACACACCT CAGCGTTG-3'
and 5'-CACGAACGTGACGAAT-3' in the presence of 2·5 mmol/L magnesium
chloride (94ºC for 8 min followed by 40 cycles of 94ºC for 1
min, 50ºC for 1 min, and 72ºC for 1 min).Coronavirus-infected fetal rhesus kidney cells were fixed in acetone and
used in an indirect immunofluorescence assay to detect a serological response
to the virus.Random RT-PCR assay To find out the genetic sequence information of an unknown RNA virus, we
did a random RT-PCR assay. Total RNA from virus-infected and virus-uninfected
fetal rhesus kidney cells were isolated. The RNA samples were reverse transcribed
with primer 5'-GCCGGAGC TCTGCAGAATTCNNNNNN-3', where N=A, T, C, or G, and
cDNA was amplified by a primer 5'-GCCGGAGCTCTGCAGAATTC-3'. Unique PCR products
(in size) in the infected cell preparation were cloned and sequenced, and
the genetic homology compared with those in GenBank.The routine receipt and inoculation of samples was done in a biosafety
level-2 laboratory. Laboratory procedures involving culture of the virus
was done in biosafety level-3 containment.Statistical analysisWe compared risk factors associated with complicated and uncomplicated
disease with the 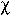 2 test for categorical variables. Continuous variables were tested by Student's
t test. A p value of less than 0·05 was taken to be significant.
We used SPSS (version 10.0) for all analyses. We did not do multivariate
analysis since the number of cases was too small for meaningful results
to be obtained.Role of the funding sourceThe sponsors of the study had no role in the study design, data collection,
data analysis, data interpretation, or in the writing of the report. 2 test for categorical variables. Continuous variables were tested by Student's
t test. A p value of less than 0·05 was taken to be significant.
We used SPSS (version 10.0) for all analyses. We did not do multivariate
analysis since the number of cases was too small for meaningful results
to be obtained.Role of the funding sourceThe sponsors of the study had no role in the study design, data collection,
data analysis, data interpretation, or in the writing of the report.

All 50 patients with SARS were ethnic Chinese. They represented five different
epidemiologically linked clusters and sporadic cases fitting the case definition.
They were admitted to hospital at a mean of 5 days (SD 2·3) after
the onset of symptoms. The median age was 42 years (range 23-74) and the
female-to-male ratio was 1 to 1·3. Among the patients, 14 (28%)
were health-care workers and five (10%) had a history of a visit to a hospital
in which there was a major outbreak of SARS, 13 (26%) were household contacts
and 12 (24%) had social contacts with patients who had SARS; four (8%)
had recently travelled to mainland China.The presenting complaint in most patients was feverishness or shortness
of breath. Cough and myalgia were present in more than half the patients
(table 1). Upper-respiratory-tract symptoms such as rhinorrhoea (n=12,
24%) and sore throat (n=10, 20%) were present in a few patients. Watery
diarrhoea (n=5, 10%) and anorexia (n=5, 10%) were also reported. At initial
examination, auscultatory findings such as crepitations and decreased air
entry were present in only 19 (38%) patients. Dry cough was reported by
31 (62%) patients. All had radiological evidence of consolidation at the
time of admission involving one zone in 36, two zones in 13, and three
zones in one.
| Clinical symptoms* |
Number (%) |
| Fever |
50 (100) |
| Chill or rigors |
37 (74) |
| Cough |
31 (62) |
| Myalgia |
27 (54) |
| Malaise |
25 (50) |
| Running nose |
12 (24) |
| Sore throat |
10 (20) |
| Shortness of breath |
10 (20) |
| Anorexia |
10 (20) |
| Diarrhoea |
5 (10) |
| Headache |
10 (20) |
| Dizziness |
6 (12) |
| *Truncal maculopapular rash was noted in one patient. |
| Table 1: Symptoms of 50 patients with SARS at presentation |
Despite high fever, 49 (98%) patients had no evidence of a leucocytosis.
In peripheral blood tests lymphopenia was present in 68%, leucopenia in
26%, thrombocytopenia in 40%, and anaemia in 18% (table 2). Alanine aminotransferase
(45-350 U/L) and creatinine kinase (141-1379 U/L) were raised in 34% and
26%, respectively.
| Laboratory variables |
Mean (range) |
Number (%) of abnormal |
Normal range |
| Haemoglobin |
12·9 (8·9-15·9) |
·· |
11·5-16·5 g/dL |
|
Anaemia |
·· |
9 (18%) |
·· |
| White-cell count |
5·17 (1·1-11·4) |
·· |
4-11x109/L |
|
Leucopenia |
·· |
13 (26%) |
·· |
| Lymphocyte count |
0·78 (0·3-1·5) |
·· |
1·5-4·0x109/L |
|
Severe lymphopenia (<1·0x109 /L) |
·· |
34 (68%) |
·· |
| Platelet count |
174 (88-351) |
·· |
150-400x109/L |
|
Thrombocytopenia |
·· |
20 (40%) |
·· |
| Alanine aminotransferase |
63 (11-350) |
·· |
6-53 U/L |
|
Raised alanine aminotransferase |
·· |
17 (34%) |
·· |
| Albumin |
37 (26-50) |
·· |
42-54 g/L |
|
Low albumin |
·· |
34 (68%) |
·· |
| Globulin |
33 (21-42) |
·· |
24-36 g/L |
|
Raised globulin |
·· |
10 (20%) |
·· |
| Creatinine kinase |
244 (31-1379) |
·· |
34-138 U/L |
|
Raised creatinine kinase |
·· |
13 (26%) |
·· |
| Table 2: Initial laboratory findings of 50 patients with SARS |
Routine microbiological investigation for known viruses and bacteria by
culture, antigen detection, and PCR was negative in most cases. Blood culture
was positive for Escherichia coli in one man aged 74 years admitted to intensive care. The finding was attributed
to a hospital-acquired urinary-tract infection. Klebsiella pneumoniae and Haemophilus influenzae were isolated from the sputum samples of two other patients on admission.Oral levofloxacin 500 mg every 24 h was given to nine patients, and amoxicillin-clavulanate
given intravenously 1·2 g at 8 h intervals or orally 375 mg three
times daily, and intravenous or oral clarithromycin 500 mg every 12 h were
given to another 40 patients. Four patients received oral oseltamivir 75
mg twice daily. In one patient, intravenous ceftriaxone 2 g every 24 h,
oral azithromycin 500 mg every 24 h, and oral amantadine 100 mg twice daily
were given for empirical coverage of typical and atypical pneumonia.At the time of writing, 19 patients had progressed to severe disease with
oxygen desaturation requiring intensive care and ventilatory support for
a mean of 6·4 days. The mean time between onset of symptoms and
worsening was 8·3 days. Intravenous ribavirin 8 mg/kg every 8 h
for 7-10 days and steroid (intravenous hydrocortisone 100 mg every 6 h,
or hydrocortisone 200 mg every 8 h, or methylprednisolone 1-3 mg/kg every
24 h for two to three doses and tailed off over 2-3 weeks) was given in
49 patients at a mean of 6·7 days after onset of symptoms. Of the
six patients given ribavirin and steroids before intubation and ventilation
in intensive care, two had a consistent response in terms of resolution
of fever, decreased respiratory support, and later radiological resolution,
whereas the other four had fluctuating fever and static requirement in
respiratory support.The risk factors associated with severe complicated disease requiring intensive
care and ventilatory support were older age, severe lymphopenia, impaired
alanine aminotransferase, and delayed starting of ribavirin and steroid
(table 3). All the complicated cases were treated with ribavirin and steroids
after admission to the intensive-care unit, whereas all the uncomplicated
cases were started on ribavirin and steroids in the general ward. As expected,
31 uncomplicated cases recovered or improved, whereas eight patients with
complicated disease worsened, with one death at the time of writing. All
50 patients had been monitored for a mean of 12 days (SD 6·1) at
the time of writing.
|
|
Complicated case (n=19) |
Uncomplicated case (n=31) |
p |
| Mean (SD) age |
49·5 (12·7) |
39·0 (10·7) |
0·005 |
| Male/female ratio |
8/11 |
14/17 |
| Underlying illness |
5* |
1õ |
0·05 |
| Method of contact |
|
Travel to China |
1 |
3 |
|
Health-care worker |
5 |
9 |
|
Hospital visit |
1 |
4 |
|
Household contact |
8 |
5 |
0·09 |
|
Social contact |
4 |
10 |
| Mean (SD) duration of symptoms to admission (days) |
5·2 (2·0) |
4·7 (2·5) |
| Mean (SD) admission temperature (C) |
38·8 (0·9) |
38·7 (0·8) |
| Mean (SD) initial total peripheral WBC count (x109/L) |
5·1 (2·4) |
5·2 (1·8) |
| Mean (SD) initial lymphocyte count (x109/L) |
0·66 (0·3) |
0·85 (0·3) |
0·04 |
| Presence of thrombocytopenia (<150x109/L) |
8 |
12 |
| Impaired liver-function test |
11 |
6 |
0·01 |
| Chest radiographic changes (number of zones affected) |
1·4 |
1·2 |
| Mean (SD) day of worsening from onset of symptomsö |
8·3 (2·6) |
Not applicable |
| Number of patients who received ribavirin and steroids |
18 |
31 |
| Mean (SD) day of start of ribavirin and steroids from onset of symptoms |
7·7 (2·9) |
5·7 (2·6) |
0·03 |
| Start of ribavirin and steroids after worsening |
12 |
0 |
0·0001 |
| Response to ribavirin and steroids |
11 |
28 |
0·02 |
| Outcome |
|
Improved or recovered |
10 |
31 |
0·0001 |
|
Not improving |
8 |
0 |
0·0004 |
|
Died |
1 |
0 |
| WBC=white-blood cell. *Two patients had diabetes mellitus, one had hypertrophic
obstructive cardiomyopathy, one had chronic active hepatitis B, and one
had brain tumour. õOne patient had essential hypertension. öDesaturation
requiring intensive-care support. Response defined as resolution of fever
within 48 h, decreased ventilatory support, or radiological improvement. |
| Table 3: Risk factors associated with severe disease requiring intensive care and
ventilatory support |
Two virus isolates, identified as a coronavirus, were isolated from two
patients. One was from an open lung biopsy sample from a male Hong Kong
Chinese resident aged 53 years and the other from a nasopharyngeal aspirate
of a woman aged 42 years with good previous health. The man had a history
of 10 h social contact with a Chinese visitor coming from Guangzhou, mainland
China, who later died from SARS. 2 days after exposure, this patient presented
with fever, malaise, myalgia, and headache. Crepitations were present over
the right lower zone and there was a corresponding alevolar shadow on the
chest radiograph. Haematological investigation revealed lymphopenia of
0·7x109/L with normal total white-cell and platelet count. Alanine aminotransferase
(41 U/L) and creatinine phosphokinase (405 U/L) were impaired. Despite
a combination of oral azithromycin, amantadine, and intravenous ceftriaxone,
there was increasing bilateral pulmonary infiltrates and progressive oxygen
desaturation. Therefore, an open lung biopsy was done 9 days after admission.
Histopathological examination showed a mild interstitial inflammation with
scattered alveolar pneumocytes showing cytomegaly, granular amphophilic
cytoplasm, and enlarged nuclei with prominent nucleoli. No cells showed
inclusions typical of herpes virus or adenovirus infection. He required
ventilation and intensive care after the surgical procedure. Empirical
intravenous ribavirin and hydrocortisone were given. He died 20 days after
admission. Coronavirus RNA was detected in his nasopharyngeal aspirate,
lung biopsy samples, and post-mortem lung samples. He had a significant
rise in antibody titre (from 1/200 to 1/1600) to his own coronavirus isolate.The female patient from whom a coronavirus was isolated had a history of
good health. She had recently travelled to Guangzhou for 2 days. She presented
with fever and diarrhoea 5 days after return to Hong Kong. Physical examination
showed crepitation over the right lower zone, which had a corresponding
alveolar shadow on chest radiograph. Investigation revealed leucopenia
(2·7x109/L), lymphopenia (0·6x109/L), and thrombocytopenia (104x109/L). Despite empirical antimicrobial coverage with amoxicillin-clavulanate,
clarithromycin, and oseltamivir, she worsened 5 days after admission and
required mechanical ventilation and intensive care for 5 days. She gradually
improved without treatment by ribavirin or steroids. Her nasopharyngeal
aspirate was positive on RT-PCR for coronavirus and she seroconverted from
titre less than one per 50 to one per 1600 to the coronavirus isolate.Viruses were isolated on fetal rhesus kidney cells from the lung biopsy
and nasopharyngeal aspirate, respectively, of these two patients. The initial
cytopathic effect noted was the appearance of rounded refractile cells
appearing 2-4 days after inoculation. The cytopathic effect did not progress
in the initial culture tubes but on subsequent passage, and appeared in
24 h. The two virus isolates did not react with the routine panel of reagents
used to identify virus isolates, including those to influenza A, B, parainfluenza
types 1, 2, and 3, adenovirus, and respiratory syncytial virus (DAKO, Glostrup,
Denmark). They also did not react in RT-PCR assays for influenza A and
human metapneumovirus, or in PCR assays for mycoplasma. The virus was ether
sensitive, which shows that it was an enveloped virus. Electron microscopy
of negative stained (3% potassium phospho-tungstate, pH 7·0) ultracentrifuged
cell-culture extracts showed the presence of pleomorphic enveloped virus
particles of around 80-90 nm (range 70-130 nm) in diameter with surface
morphology compatible with a coronavirus (figure 1). Thin-section electron
microscopy of infected cells revealed virus particles of 55-90 nm diameter
within smooth walled vesicles in the cytoplasm (figure 2, B). Virus particles
were also seen at the cell surface. The overall findings were compatible
with coronavirus infection in the cells.
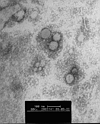 |
| Figure 1: Electron microscopy of ultracentrifuged deposit of cell-culture-grown human
pneumonia-associated coronavirusNegatively stained with 3% potassium phospho-tungstate, pH 7·0. |
 |
| Figure 2: Thin-section electron micrograph of lung biopsy sample from patient with
SARS (A) and of human pneumonia-associated coronavirus infected cells (B)
|
A thin-section electron micrograph of the lung biopsy sample from the 53-year-old
male contained 60-90 nm viral particles in the cytoplasm of desquamated
cells. These viral particles were similar in size and morphology to those
observed in the cell cultured virus isolate from both patients (figure
2, A).The RT-PCR products generated in a random primer RT-PCR assay were analysed,
and unique bands found in the virus-infected samples were cloned and sequenced.
Of 30 clones examined, one containing 646 bp of unknown origin was identified.
Sequence analysis of this DNA fragment suggested this sequence had a weak
homology to viruses of the family of Coronaviridae. Deducted aminoacid
sequence (215 aminoacids) from this unknown sequence, however, had the
highest homology (57%) to the RNA polymerase of bovine coronavirus and
murine hepatitis virus, confirming that this virus belongs to the family
of Coronaviridae. Phylogenetic analysis of the protein sequences showed
that this virus, although most closely related to the group II coronaviruses,
was a distinct virus (figure 3).
 |
| Figure 3: Phylogenetic analysis of the partial protein sequence (215 aminoacids)
of the coronavirus (SARS)GenBank accession number AY268070. Tree is constructed by neighbour-jointing
method. Horizontal-line distance represents number of sites at which the
two sequences compared are different. Bootstrap values deducted from 500
replicates. |
Based on the 646 bp sequence of the isolate, specific primers for detecting
the new virus were designed for RT-PCR detection of this human pneumonia-associated
coronavirus genome in clinical samples. Of the 44 nasopharyngeal samples
available from the 50 SARS patients, 22 had evidence of human pneumonia-associated
coronavirus RNA. Viral RNA was detectable in ten of 18 faecal samples tested.
The specificity of the RT-PCR reaction was confirmed by sequencing selected
positive RT-PCR-amplified products. None of 40 nasophararyngeal and faecal
samples from patients with unrelated diseases were reactive on RT-PCR.In 35 of the 50 most recent serum samples from patients with SARS there
was evidence of antibody to the coronavirus. Of 32 patients from whom paired
acute and convalescent sera were available, all had seroconverted or had
more than a four-fold increase in antibody titre to the virus. Five other
pairs of sera from additional SARS patients from clusters outside this
study group were also tested to provide a wider sampling of SARS patients
in the community and all of them seroconverted. None of 80 sera from patients
with respiratory or other diseases and none of 200 blood donors had detectable
antibody.If seropositivity to human pneumonia-associated coronavirus in one serum
sample or viral RNA detection in the nasopharyngeal aspirates or stools
is deemed evidence of infection with the coronavirus, 45 of the 50 patients
have evidence of infection. Of the five patients with no virological evidence
of coronavirus infection, only one had a serum sample tested more than
14 days after onset of clinical disease.

The outbreak of SARS is unusual in several ways, especially in the appearance
of clusters of patients with pneumonia in health-care workers and family
contacts. In this series of patients, investigations for conventional pathogens
of atypical pneumonia proved negative. However, a virus belonging to the
family Coronaviridae was isolated from the lung biopsy and nasopharyngeal
aspirate of two SARS patients and other patients with SARS had a serological
response to this virus.The family Coronaviridae includes the genus Coronavirus and Torovirus.
They are enveloped RNA viruses that cause disease in human beings and animals.
The previously known human coronaviruses, types 229E and OC43 are a major
cause of the common cold.6 They can occasionally cause pneumonia in older adults, neonates, or immunocompromised
patients.7,8 Coronaviruses have been reported to be an important cause of pneumonia
in military recruits, accounting for up to 30% of cases in some studies.9 Human coronaviruses can infect neurons, and viral RNA has been detected
in the brain of patients with multiple sclerosis.10 On the other hand, several animal coronaviruses (eg, porcine transmissible
gastroenteritis virus, murine hepatitis virus, and avian infectious bronchititis
virus) cause respiratory, gastrointestinal, neurological, or hepatic disease
in their respective hosts.11 Phylogenetically, human pneumonia-associated coronavirus was not closely
related to any known human or animal coronavirus or torovirus. We based
our analysis on a 646 bp fragment of the polymerase gene which showed that
the virus belongs to antigenic group 2 of the coronaviruses, along with
murine hepatitis virus and bovine coronavirus. However, viruses of the
Coronaviridae can undergo heterologous recombination within the virus family
and genetic analysis of other parts of the genome needs to be done before
the nature of this new virus is more conclusively defined.6 The biological, genetic, and clinical data taken together show that the
new virus is not one of the two known human coronaviruses. Antibody to
the previously recognised human 229E and OC43-like coronaviruses is widespread
in the human population.12 The lack of serological reactivity against the novel pneumonia-associated
coronavirus among our patients implies that there is little antigenic cross
reactivity between it and the 229E or OC43 viruses.Most patients who had clinically defined SARS had either serological or
RT-PCR evidence of infection by this virus. By contrast, neither antibody
nor viral RNA was detectable in healthy controls. All 32 patients from
whom acute and convalescent sera were available had rising antibody titres
to human pneumonia-associated coronavirus, which strengthens the contention
that a recent infection with this virus is a necessary factor in the evolution
of SARS. In addition, all five pairs of acute and convalescent sera tested
from patients from other hospitals in Hong Kong also showed seroconversion
to the virus. Five patients who had SARS had no serological or virological
evidence of coronavirus infection. They need to have later convalescent
sera tested to define whether they seroconvert subsequently. However, the
concordance of human pneumonia-associated coronavirus with the clinical
definition of SARS seems remarkable, given that clinical case definitions
are never perfect.No evidence of human-metapneumovirus infection, by RT-PCR or rising antibody
titre, was detected in any of our patients and no other pathogen was consistently
detected. It is therefore highly likely that that this coronavirus is either
the cause of SARS or a necessary prerequisite for disease progression.
Whether other microbial or non-microbial cofactors play a part in progression
of the disease remains to be investigated.We describe the clinical presentation and complications of SARS. Less than
25% of patients with coronaviral pneumonia had upper-respiratory-tract
symptoms. As expected in atypical pneumonia, both respiratory symptoms
and positive auscultatory findings were disproportionally mild compared
with the chest radiographic findings. Gastrointestinal symptoms were present
in 10% of patients. These symptoms are relevant since the viral RNA is
detectable in faeces of some patients and coronaviruses have been associated
with diarrhoea in animals and human beings.13 The high incidence of altered liver function, leucopenia, severe lymphopenia,
thrombocytopenia, and subsequent evolution into adult respiratory distress
syndrome suggests a severe systemic inflammatory damage induced by this
human pneumonia-associated coronavirus. Thus immunomodulation by steroid
treatment may be important to complement the empirical antiviral treatment
with ribavirin. It is pertinent that severe human disease associated with
the avian influenza subtype H5N1, another virus that has crossed from animals
to human beings, has also been postulated to have an immunopathological
component.14 In common with H5N1 disease, patients with severe SARS are adults, have
lymphopenia, and have variables of organ dysfunction beyond the respiratory
tract.15 A window of opportunity of around 8 days exists from the onset of symptoms
to respiratory failure. Severe complicated cases are strongly associated
with underlying disease and delayed use of ribavirin and steroid treatment.
After our clinical experience in the first cases, we started this combination
treatment very early in subsequent cases, which were generally uncomplicated
at the time of admission. The overall mortality at the time of writing
was only 2% with use of this treatment regimen. Eight of 19 complicated
cases still had shown no notable response. A detailed analysis of the therapeutic
response to this combination regimen is impossible, given the heterogeneous
dosing and time of starting treatment. The choice of ribavirin was made
on empirical grounds, before the cause was identified. It might have to
be reviewed once the in-vitro susceptibility of human pneumonia-associated
coronavirus to antivirals is better understood.Another factor associated with severe disease is acquisition of the disease
through household contact. People infected in this way may have a higher
dose or duration of viral exposure and the presence of underlying diseases
than people exposed, for example, through social contact.Our clinical description pertains largely to the more severe cases admitted
to hospital. We presently have no data on the full clinical spectrum of
the emerging coronavirus infection in the community or among outpatients.
The availability of diagnostic tests we describe will help address these
questions. In addition, it will allow questions to be addressed about the
period of virus shedding (and communicability) during convalescence, the
presence of virus in other body fluids and excreta, and the presence of
virus shedding during the incubation period.The epidemiological data at present seem to suggest that the virus is spread
by droplets or by direct and indirect contact, although airborne spread
cannot be ruled out. The finding of infectious virus in the respiratory
tract supports this contention. Preliminary evidence also suggests that
the virus may be shed in the faeces. However, detection of viral RNA does
not prove that the virus is viable or transmissible. If viable virus is
detectable in the faeces, this is potentially an additional route of transmission.
Several animal coronaviruses are spread via the faecal-oral route.11 Samples from patients with SARS were not readily distinguishable from
samples from other patients on receipt in the laboratory. Thus, initial
processing of these samples was done under biohazard level-2 containment.
However, culture of the virus was done in biosafety level-3 containment.
These containment measures have proved successful so far, in that no laboratory
infections have been documented.We have provided evidence that a virus in the coronavirus family is the
causal agent of SARS. However it remains possible that other viruses act
as opportunistic secondary invaders to increase the disease progression,
a hypothesis that needs to be investigated further.
J S M Peiris and K Y Yuen are co-principal investigators, jointly wrote
the report, and supervised the virological and clinical components of the
study. L L M Poon obtained viral sequence data and developed the RT-PCR
assay. Y Guan, K H Chan, W Lim, and J M Nicholls did the phylogenetic analysis,
clinical virology, and electron microscopy to identify the novel virus.
T K Ng, D N C Tsang, R Yung, and W Lim coordinated the microbiological
investigations and analysed the overall results. All other researchers
were involved in collection and analysis of the clinical data.
| Members of the HKU SARS study group |
I F N Hung, Department of Medicine, S W Kwan, K F Lo, W H Seto, Department
of Microbiology, Queen Mary Hospital, Hong Kong; O T Y Tsang, E Y K Tso,
Department of Medicine, Princess Margaret Hospital; Queen Mary Hospital,
Hong Kong; W Luk, H Y Ng, L J Zhang, C Y Cheung, O K Wong, W Cheung, Department
of Microbiology, University of Hong Kong.
| Conflict of interest statement |
None declared.
We thank E K Yeoh of the Hong Kong Government for facilitating this study.
This work was greatly facilitated by the WHO Network of laboratories investigating
the causes of SARS. The laboratories are Centers for Disease Control and
Prevention, Atlanta, GA, USA; The Chinese University of Hong Kong; Erasmus
Universiteit, Rotterdam, Netherlands; The Government Virus Unit, Hong Kong
SAR; Institut für Medizinische Virologie im Klinikum der Johann Wolfgang
Goethe-Universität Frankfurt am Main, Frankfurt, Germany; Institut
Pasteur, Paris, France; National Institute of Infectious Diseases, Tokyo,
Japan; National Microbiology Laboratory, Population Public Health Branch,
Health Canada; Public Health Laboratory Service, Colindale, London, UK;
The University of Hong Kong, Virology Unit; Singapore General Hospital,
Singapore. We thank the staff of the Department of Microbiology, Queen
Mary Hospital, Hong Kong, and the Government Virus Unit of the Department
of Health for their technical assistance. We received research funding
from Public Health Research (grant A195357), the National Institute of
Allergy and Infectious Diseases, USA, the University of Hong Kong, and
the Hospital Authority of Hong Kong SAR.

1 WHO. Severe acute respiratory syndrome (SARS). Wkly Epidemiol Rec 2003; 78: 86.2 WHO. Severe acute respiratory syndrome (SARS). Wkly Epidemiol Rec 2003; 78: 81-83. [PubMed]3 Chan KH, Maldeis N, Pope W, et al. Evaluation of directigen flu A+B test
for rapid diagnosis of influenza A and B virus infections. J Clin Microbiol 2002; 40: 1675-80. [PubMed]4 Wiedbrauk DL, Johnston SLG. Manual of clinical virology. New York: Raven
Press, 1993.5 Fouchier RA, Bestebroer TM, Herfst S, Van Der Kemp L, Rimmelzwaan GF,
Osterhaus AD. Detection of influenza A virus from different species by
PCR amplification of conserved sequences in the matrix gene. J Clin Microbiol 2000; 38: 4096-101. [PubMed]6 Holmes KV. Coronaviruses. In: Knipe DM, Howley PM, eds. Fields Virology,
4th edn. Philadelphia: Lippincott Williams and Wilkins, 2001: 1187-203.7 El-Sahly HM, Atmar RL, Glezen WP, Greenberg SB. Spectrum of clinical
illness in hospitalized patients with "common cold" virus infections.
Clin Infect Dis 2000; 31: 96-100. [PubMed]8 Fotz RJ, Elkordy MA. Coronavirus pneumonia following autologous bone
marrow transplantation for breast cancer. Chest 1999; 115: 901-05. [PubMed]9 Wenzel RP, Hendley JO, Davies JA, Gwaltney JM Jr. Coronavirus infections
in military recruits: three-year study with coronavirus strains OC43 and
229E . Am Rev Respir Dis 1974; 109: 621-24. [PubMed]10 Talbot PJ, Cote G, Arbour N. Human coronavirus OC43 and 229E persistence
in neural cell cultures and human brains. Adv Exp Med Biol (in press).11 McIntosh K. Coronaviruses: a comparative review. Curr Top Microbiol Immunol 1974; 63: 85-129. [PubMed]12 McIntosh K, Kapikian AZ, Turner HC, Hartley JW, Parrott RH, Chanock
RM. Seroepidemiologic studies of coronavirus infection in adults and children.
Am J Epidemiol 1970; 91: 585-92. [PubMed]13 Caul EO, Egglestone SI. Further studies on human enteric coronaviruses
Arch Virol 1977; 54: 107-17. [PubMed]14 Cheung CY, Poon LLM, Lau ASY, et al. Induction of proinflammatory cytokines
in human macrophages by influenza A (H5N1) viruses: a mechanism for the
unusual severity of human disease. Lancet 2002; 360: 1831-37. [Text]15 Yuen KY, Chan PKS, Peiris M, et al. Clinical features and rapid viral
diagnosis of human disease associated with avian influenza A H5N1 virus.
Lancet 1998; 351: 467-71. [Text]

|